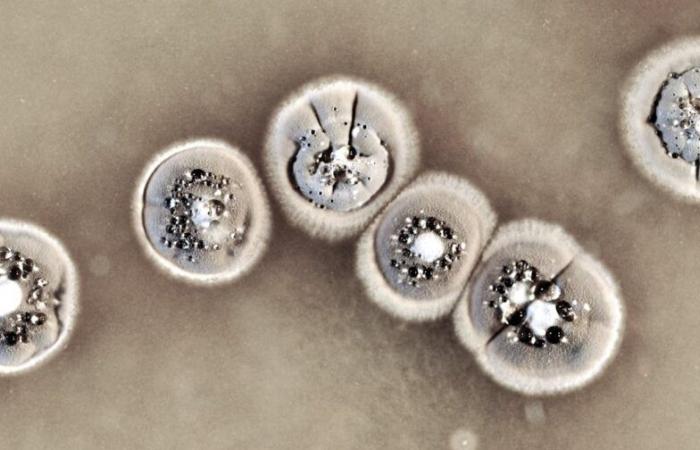
Certain bacteria are capable of producing antibiotics in a context of competition between species, this is the case of the genus Streptomyces. A new discovery dissects the way they produce these molecules and opens the way to the production of new antibiotics and the fight against antibiotic resistance.
The spread of antibiotic resistance among human and animal pathogens is at the origin of a contemporary health crisis and a potential future catastrophe. The World Health Organization (WHO) recognizes antimicrobial resistance as a major socio-economic problem and “one of the 10 greatest threats to global public health”. It is now accepted that the massive use and misuse of antibiotics favors the emergence and spread of antimicrobial resistance in human pathogens. A real speed race is underway: it is imperative to innovate to identify and produce new molecules capable of countering the adaptation of bacteria to antibiotic pressure.
The main source of natural molecules for antibiotic use are soil bacteria, particularly Streptomyces. In fact, bacteria establish, thanks to the molecules they synthesize, a balance with other organisms colonizing the soil, including competition which consists of eliminating the competition.
Nearly a third of all known antibiotics originate from Streptomyces, which places this single bacterial genus in second position for the discovery of antibiotics, just behind all higher plants.
When bacteria play chemists
Streptomyces are non-pathogenic bacteria that are among nature’s most skilled “chemists” thanks to their ability to produce a wide diversity of specialized metabolites. These metabolites are used in human and veterinary medicine as well as in agronomy, including antibiotic, antiproliferative, antitumor or antioxidant molecules. For example, streptomycin produced by the bacteria Streptomyces griseuswas discovered as an effective antibiotic against tuberculosis by Selman Waksman, winner of the Nobel Prize in Medicine in 1952.
After the golden age of the discovery of antibiotics in the 1960s which anticipated the end of infectious diseases, the appearance of resistant strains was not accompanied by the discovery and marketing of new therapeutic agents. The fault lies in the high probability of reisolating an already known molecule several times. Besides the fact that the extent of the diversity of molecules produced is not known, the processes (mechanisms and pressures) favoring the diversity of these molecules in producing organisms are largely unknown. Can we hope to benefit from a renewal of our antibiotic arsenal through the ability of producers to diversify their biosynthesis genes?
Over the past decade, advances in sequencing technologies have revealed that Streptomyces genomes are full of unsuspected potential for the synthesis of metabolites of interest. Indeed, while each strain was known most often for the synthesis of a molecule revealed by its activity, their genome includes up to 40 groups of biosynthesis genes (biosynthetic gene clustersBCG) representing a total of up to 10% of their genetic heritage. These are the organizations that dedicate the majority of their assets to this function. Each of these BGCs theoretically allows the synthesis of a new molecule. The era of genomics had just opened with its hopes and its frustrations: hopes of a new diversity to explore, frustrations of not mastering the processes of creating diversity.
The ground, a real battlefield
The richness and diversity of antibiotics produced by Streptomyces can be fundamentally explained by their ecological role for the bacteria in its ecosystem. Streptomyces live mainly in soils and particularly in the rhizosphere, the zone of the soil under the influence of the plant’s roots, where the plant microbiota develops. This heterogeneous ecosystem with changing physicochemical conditions is the site of fierce competition for nutrients, but also of fertile symbioses.
Streptomyces are involved in these multiple and complex biotic interactions with other organisms living in the soil (rhizospheric bacteria, fungi, plants and animals) through the metabolites produced that they synthesize and excrete (constituting the interactome Streptomyces). These chemical dialogues allow the balance of communities living in the soil.
Under this strong biological pressure, the arms race is permanent under our feet to ensure the production of new active molecules guaranteeing the survival and dissemination of the bacteria. Resistance to the drug synthesized by the producer himself (in order to avoid suicide) must evolve as a mirror of the biosynthesis capacity. We have shown that within a population of Streptomyces, while a single strain produces the antibiotic, the non-producing parent strains are resistant, thus favoring the dissemination of the producer and its relatives. In doing so, the producers are also a reservoir of resistance in the making (we speak of “proto-resistance”, ancestor of the resistance identified in our pathogens).
Colossal genetic exchanges
At the DynAMic laboratory, we have recently shown that Streptomyces are capable of exchanging large quantities of DNA when in contact with each other. Indeed, these gene transfers or horizontal transfer occur during conjugation between strains originating from the rhizosphere. The Streptomyces genome is rich in mobile genetic elements called AICE, to actinomycetes integrative and conjugative elements (conjugative and integrative element of actinomycetes), which promote, in addition to the mobility of their own genomic DNA (between 20 and 40 kilobases), that of chromosomal DNA between the partners involved in the crossing.
During a single cross, the transfer of one or more of these AICEs is accompanied by that of several DNA fragments representing a total of up to 30% of the genetic heritage of the donor parent.
Provided by the author
As the genome of the Streptomyces used in this experiment includes approximately 12 million base pairs, this represents several hundred or even thousands of kilobases, and therefore as many genes (1 gene ≃ 1,000 bases in bacteria)! Enough for entire biosynthetic pathways to be transferred; the genes necessary for the synthesis and resistance to the drug produced being grouped into regions of approximately a hundred kilobases.
Once in the recipient parent, the donor’s DNA inserts and replaces the information on the chromosome and brings about genetic innovations. Thus, we have demonstrated that more than 90% of the descendants obtained between an AICE donor bacteria and a recipient present an arsenal of metabolite biosynthesis distinct from the two parents; descendants gained or lost entire biosynthetic pathways, or combined two pathways to generate a new one.
This new phenomenon ensures genetic mixing, which can be compared to that occurring during meiosis (cell division allowing the formation of gametes) of organisms with sexual reproduction. This mixing will induce drastic alterations in specialized metabolism leading to new variants of antibiotics which would facilitate the adaptation of Streptomyces populations to their ecosystem. The rearrangement of BGCs could play a role in intra- and interspecific “bacterial warfare”. On longer time scales, it is this pressure that could promote the divergence of bacterial species by specializing them towards a specific ecosystem. It is known that genetic recombination considerably influences the process of divergence or speciation.
This discovery opens new perspectives for biotechnology and medicine. In addition to providing a better understanding of the mechanisms of DNA transfer and recombination in order to better understand the evolution and adaptation of soil bacteria, it makes it possible to exploit this phenomenon to diversify the genes and groups of genes of biosynthesis of specialized metabolites. This facilitates the discovery of new biomolecules and could accelerate the fight against bacterial resistance by providing a constant source of new antibiotics. Evolutionary tinkering, dear to François Jacob, surprises us by constantly manufacturing something new from what already exists, thus offering a constantly renewed opportunity to discover new bioactive compounds, including antibiotics, essential in the war against pathogens.